Biography
Simon Renny-Byfield is a postdoc in the Department of Plant Sciences at the University of California, Davis. He currently works on the population genetics of copy-number variants in a wild population of maize. In a past life, Simon studied the evolution of expression following gene duplication, the evolution of gene expression networks and the evolution of repetitive DNA in allopolyploid plant hybirds. He is passionate about using computational techniques to understand the diversity of plant life.
Simon started his career in science as an undergrad at Queen Mary University of London, where he studied genetics, with a particular focus on polyploidy in plants. His first paper evolved out of work done as an undergrad in the lab of Professor Andrew Leitch. In 2012, Simon earned his PhD in Plant Genomics and Computational Biology at the University of London, UK. Next was a trip over the Atlantic to the lab of Professor Jonathan F Wendel in the Ecology, Evolution and Organismal Biology department at Iowa State. More recently, Simon has been working with the award winning genius Dr Jeff Ross-Ibarra at the Department of Plant Science at the University of California, Davis.
click here for my CV, google scholar profile and twitter
Follow @TwitterDev
research highlights
My Co-Author Network
I generated this graph with some custom code using perl, R and shiny.
The graph is generated using a bibtex file containing all my publications. Using custom perl scripts I extract the number of papers each unique pair of co-authors share. With this information I use the R package igraph to generate a graph object detailing the relationships between authors. Following this, I use the R package shiny to generate the reactive graph. It should be fairly easy to see that there are really two “groups” of authors, one european and on from the USA (from my PhD and first post-doc respectively). The code will soon be available on github.
Genetic Drift Simulations
Check out the simple genetic drift simulator. It is written using R and Shiny. It is remarkably straight forward to make these reactive plots using R.
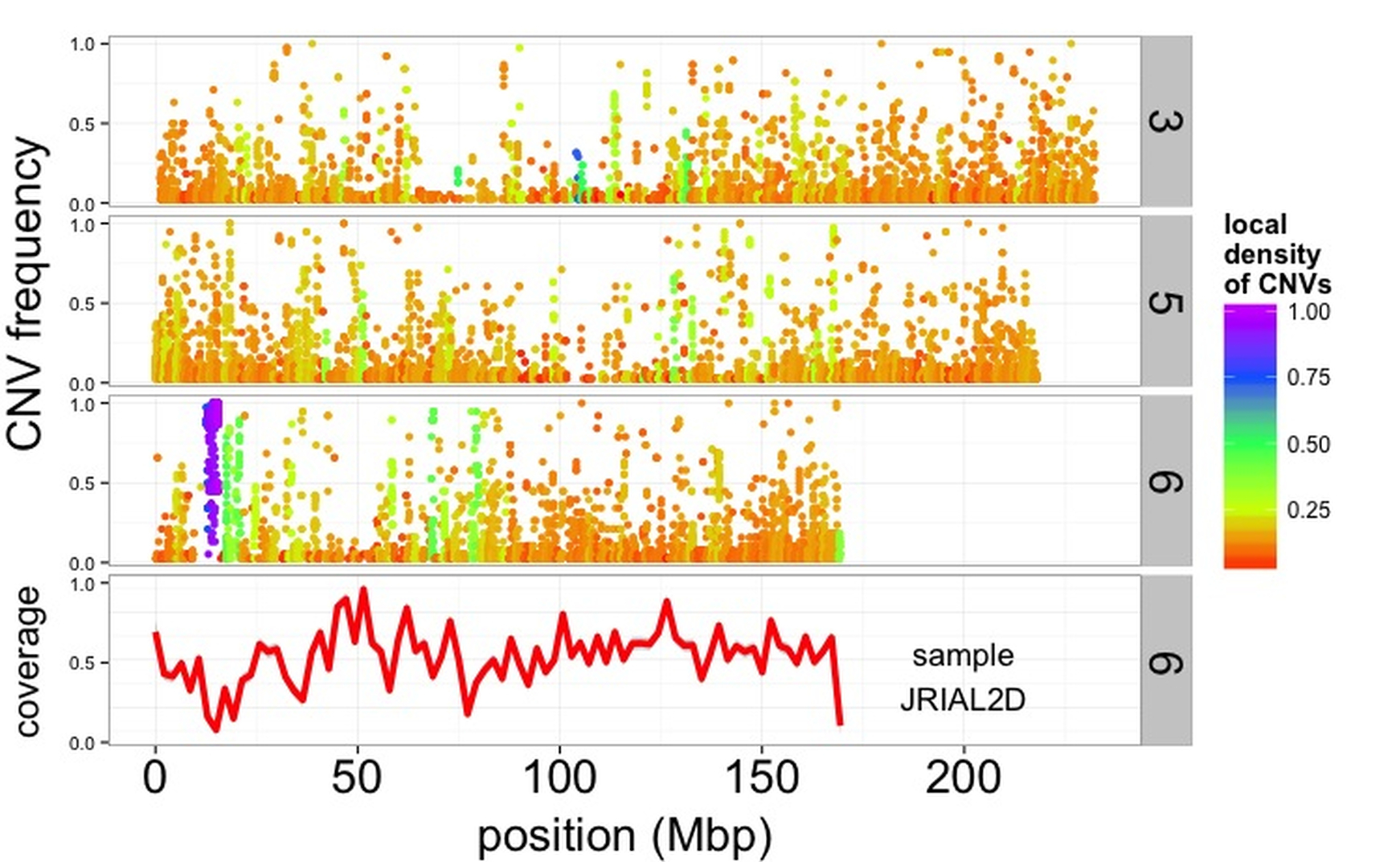
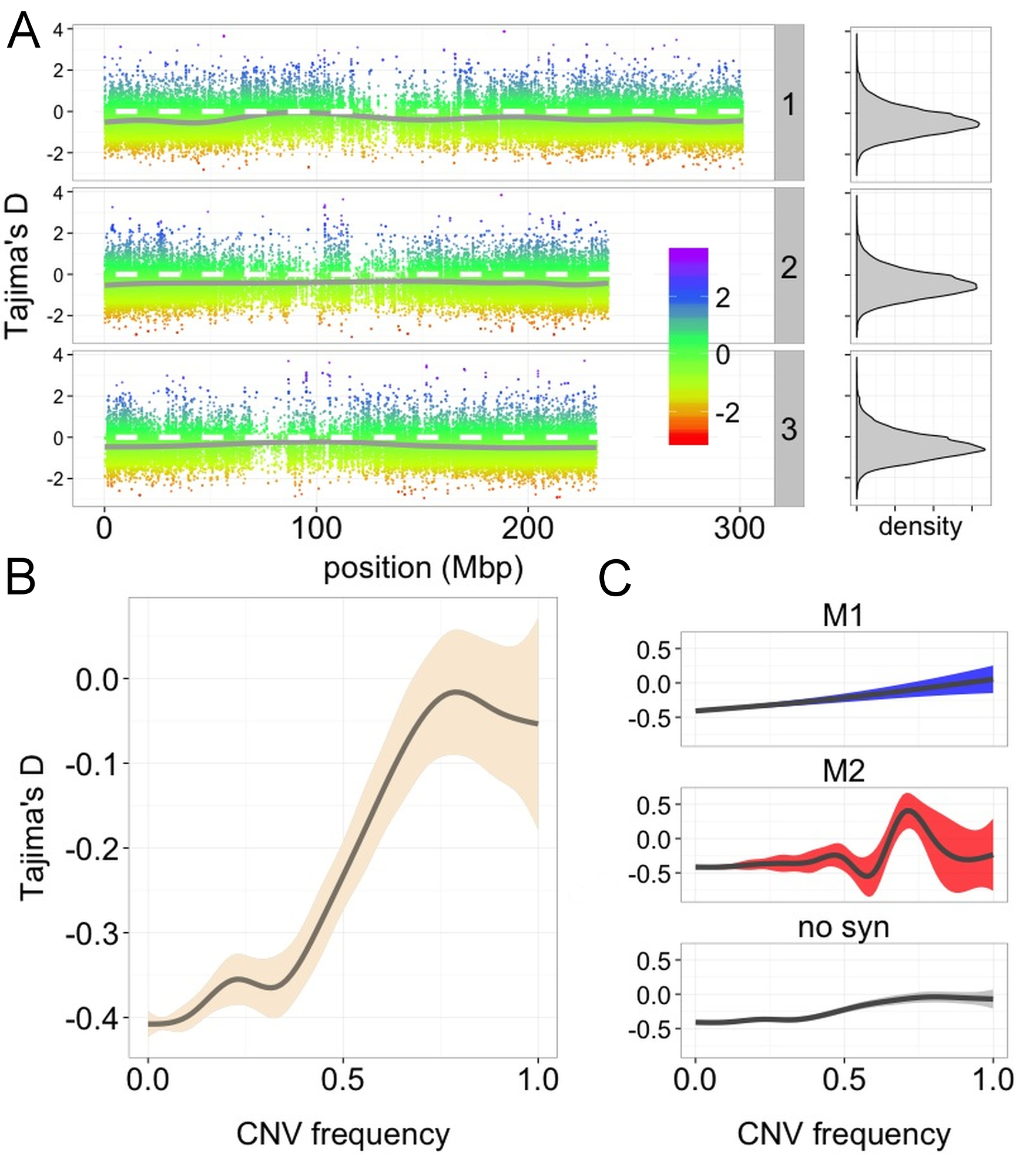
Copy-number variants (CNVs) in wild maize
I am currently developing a novel CNV calling pipeline, currently in testing. Preliminary data (see figures below) suggest that the pipeline is working well and we have identified a ~10 MB deletion on chromosome 6 segregating in a wild population of maize (otherwise known as teosinte). Furthermore we note that estimates of Tajima’s D over regions where CNV segregate differ from the background “genome-wide” level.
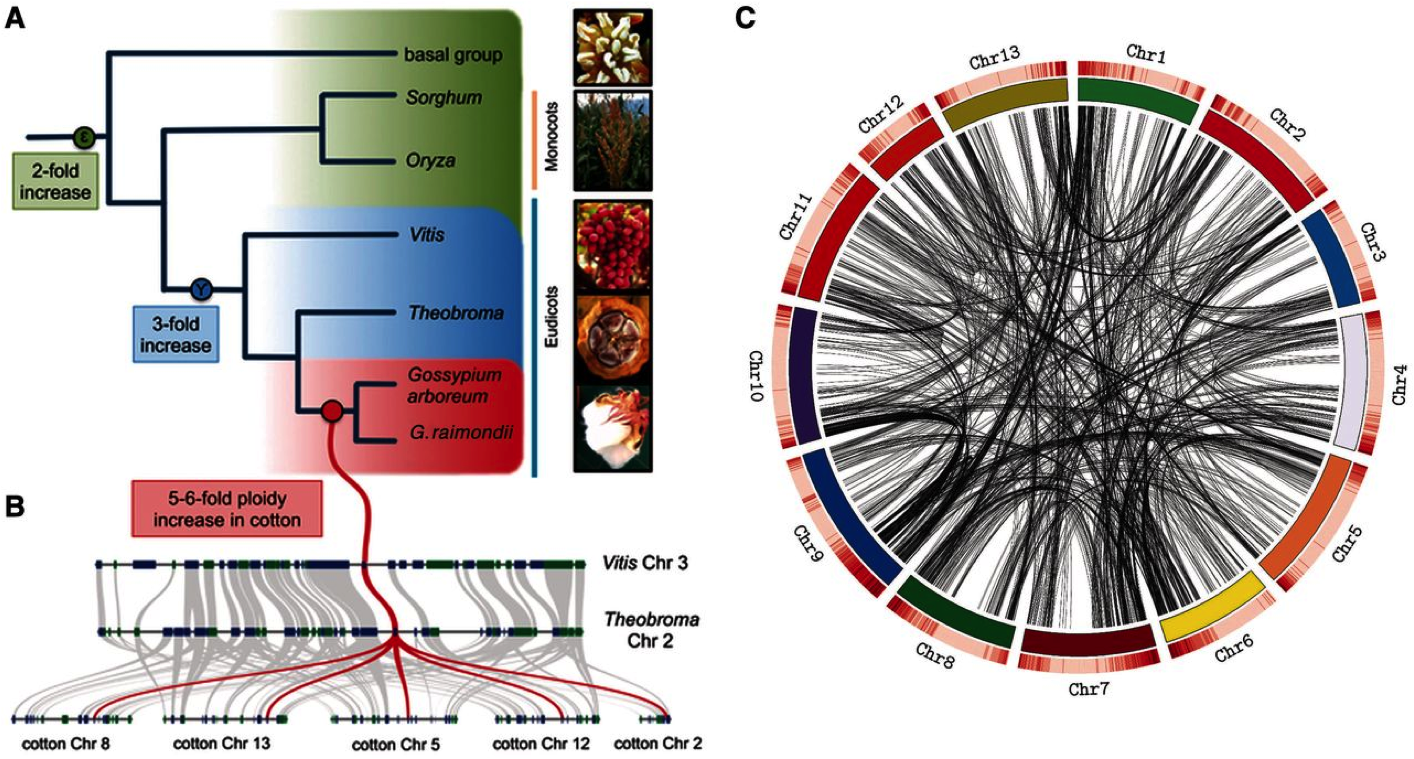
The ancient sub-genomes of cotton
Previous work has examined gene loss following ancient whole genome duplication (WGD; sometimes called polyploidy) in the cotton lineage. I showed that gene loss (sometimes called fractionation) following WGD is ongoing and bias between the duplicate sub-genomes. I further suggested that this process is the result of differing recombination rates between sub-genomes. Read the full story here.
Repetitive DNA loss in tobacco
My PhD work focused on the evolution of repetitive DNA in tobacco. Mostly the work focused on the use of fluorescent in situ hybridisation (FISH) and a custom build of an NGS clustering algorithm called RepeatExplorer which identifies and quantifies repeatitive sequences in complex genomes.
publications
In peer reviewed journals
2015
Gong, L., R. E. Masonbrink, C. E. Grover, S. Renny-Byfield and J. F. Wendel (2015). “A Cluster of Recently Inserted Transposable Elements Associated with siRNAs in.” The Plant Genome. 8 (2)
Renny-Byfield, S., L. Gong, J. P. Gallagher and J. F. Wendel (2015). “Persistence of Subgenomes in Paleopolyploid Cotton after 60 My of Evolution.” Molecular Biology and Evolution 32(4):1063-1071.
Masonbrink, R. E., J. P. Gallagher, J. J. Jareczek, S. Renny-Byfield, C. E. Grover, L. Gong and J. F. Wendel (2014). “CenH3 evolution in diploids and polyploids of three angiosperm genera.” BMC Plant Biology 14(1): 383.
2014
Renny-Byfield, S., J. P. Gallagher, C. E. Grover, E. Szadkowski, J. T. Page, J. A. Udall, X. Wang, A. H. Paterson and J. F. Wendel (2014). “Ancient Gene Duplicates in Gossypium (Cotton) Exhibit Near-Complete Expression Divergence.” Genome Biology and Evolution 6(3): 559-571.
Renny-Byfield, S. and J. F. Wendel (2014). “Doubling down on genomes: Polyploidy and crop plants.” American Journal of Botany 101(10): 1-15.
2013
Renny-Byfield, S., A. Kovarik, L. J. Kelly, J. Macas, P. Novak, M. W. Chase, R. A. Nichols, M. R. Pancholi, M.-A. Grandbastien and A. R. Leitch (2013). “Diploidization and genome size change in allopolyploids is associated with differential dynamics of low- and high-copy sequences.” The Plant Journal 74(5): 829-839.
2012
Buggs, R. J., S. Renny-Byfield, M. Chester, I. E. Jordan-Thaden, L. F. Viccini, S. Chamala, A. R. Leitch, P. S. Schnable, W. B. Barbazuk, P. S. Soltis and D. E. Soltis (2012). “Next-generation sequencing and genome evolution in allopolyploids.” American Journal of Botany 99: 372-382.
Kelly, L. J., A. R. Leitch, M. F. Fay, S. Renny-Byfield, J. Pellicer, J. Macas and I. J. Leitch (2012). “Why size really matters when sequencing plant genomes.” Plant Ecology and Diversity 5(4): 415-425.
Matyasek, R., S. Renny-Byfield, J. Fulnecek, J. Macas, M.-A. Grandbastien, R. Nichols, A. Leitch and A. Kovarik (2012). “Next generation sequencing analysis reveals a relationship between rDNA unit diversity and locus number in Nicotiana diploids.” BMC Genomics 13: 722.
Renny-Byfield, S. (2012). Evolution of repetitive DNA in angiosperms: Examples from Nicotiana allopolyploids, University of London.
Renny-Byfield, S., A. Kovarik, M. Chester, R. A. Nichols, J. Macas, P. Novak and A. R. Leitch (2012). “Independent, rapid and targeted loss of a highly repetitive DNA sequence derived from the paternal genome donor in natural and synthetic Nicotiana tabacum.” PLoS One 7(5): e36963.
2011
Kovarik, A., S. Renny-Byfield and A. R. Leitch (2011). Evolutionary implications of genome and karyotype restructuring in Nicotiana tabacum. L. Polyploidy and Genome Evolution. P. S. Soltis and D. E. Soltis. New York, Springer: 209-224.
Renny-Byfield, S., M. Chester, A. Kovařík, S. C. Le Comber, M.-A. Grandbastien, M. Deloger, R. A. Nichols, J. Macas, P. Novák, M. W. Chase and A. R. Leitch (2011). “Next generation sequencing reveals genome downsizing in allotetraploid Nicotiana tabacum, predominantly through the elimination of paternally derived repetitive DNAs.” Molecular Biology and Evolution 28(10): 2843-2854.
2010
Koukalova, B., A. P. Moraes, S. Renny-Byfield, R. Matyasek, A. R. Leitch and A. Kovarik (2010). “Fall and rise of satellite repeats in allopolyploids of Nicotiana over c. 5 million years.” New Phytologist 186(1): 148-160.
Renny-Byfield, S., M. Ainouche, I. J. Leitch, K. Y. Lim, S. C. Le Comber and A. R. Leitch (2010). “Flow cytometry and GISH reveal mixed ploidy populations and Spartina nonaploids with genomes of S. alterniflora and S. maritima origin.” Annals of Botany 105(4): 527-533.
Submitted
…it takes time to science!!!
In prep
Awards
-
2014 Irene Manton Prize for the best thesis in Botany from the Linnean Society of London.
-
2008 Bevan Prize from Queen Mary University of London, for outstanding academic achievement in Genetics.